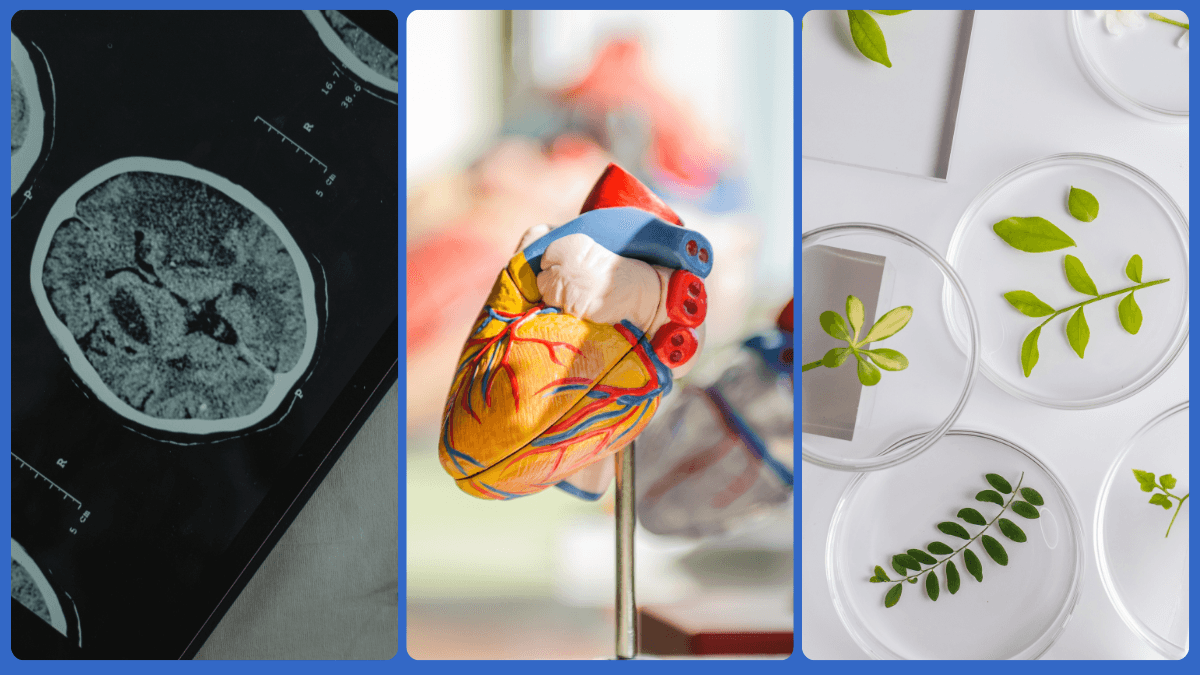
RNA sequencing (RNA-seq) emerged from the advent of NGS technologies in the mid-2000s. It provided a snapshot of the transcriptome at a specific moment, enabling researchers to investigate a cell’s function, state, and response to its environment at that time. Crucially, it also operated at resolutions orders of magnitude greater than whole-tissue analysis. Single nuclei RNA sequencing (snRNA-seq) specifically, provided unprecedented levels of detail. It allowed researchers to discover new cell types, understand complex tissue architectures, and gain truly novel insights into diseases.
1. Decoding Brain Complexity and Development
The human brain is one of the most intricate organs, comprising a vast array of cell types with distinct functions and developmental pathways. snRNA-seq allows researchers to dissect this complexity by profiling the gene expression of individual neuronal and glial nuclei. This detailed mapping is crucial for understanding brain development, neuronal differentiation, and the cellular mechanisms underlying neurological disorders.
For example, researchers have utilized snRNA-seq to study the developing human cortex. They have identified distinct cell types and uncovered the temporal dynamics of gene expression during neurogenesis. These insights are pivotal for understanding developmental disorders like autism and intellectual disabilities and can inform the development of targeted therapeutic interventions.
2. Advancing Cancer Research and Precision Oncology
Cancer is characterized by cellular heterogeneity, with tumor cells exhibiting diverse genetic and phenotypic profiles. snRNA-seq provides a granular view of this heterogeneity. It enables the identification of rare cell populations that may drive tumor progression, metastasis, or resistance to therapy. By analyzing individual nuclei from tumor samples, researchers can:
- Delineate the cellular architecture of tumors
- Identify novel biomarkers
- Uncover the molecular signatures of cancer stem cells.
In breast cancer research, for instance, snRNA-seq has been employed to reveal the presence of subpopulations of cancer cells with unique gene expression patterns associated with aggressive phenotypes. This information is invaluable for developing personalized treatment strategies and advancing precision oncology.
3. Unraveling the Mysteries of Heart Disease
Heart disease remains a leading cause of morbidity and mortality worldwide. Understanding the cellular and molecular mechanisms underlying cardiac dysfunction is crucial for developing effective therapies. SnRNA-seq is valuable for studying heart disease by enabling the analysis of gene expression in individual cardiac nuclei.
Researchers have applied snRNA-seq to:
- Investigate the cellular responses to myocardial infarction
- Identify changes in gene expression
- Signal pathways in various cardiac cell types.
These studies have highlighted potential therapeutic targets and provided insights into the regenerative capacity of the heart, opening new avenues for regenerative medicine approaches.
4. Exploring Immune System Dynamics
The immune system is a dynamic network of cells that orchestrate complex responses to pathogens and maintain homeostasis. SnRNA-seq allows researchers to study immune cells at an unprecedented resolution, providing insights into their development, differentiation, and function.
In the context of infectious diseases, snRNA-seq can profile immune responses in tissues affected by pathogens. These include the influenza virus or Mycobacterium tuberculosis. Through analyzing individual immune cell nuclei, researchers can:
- Pinpoint cellular subsets that play key roles in the immune response
- Uncover mechanisms of immune evasion by pathogens
- Inform the design of vaccines and immunotherapies.
5. Investigating Plant Biology and Crop Improvement
Beyond its applications in human health, snRNA-seq is also transforming plant biology. It can enable the study of gene expression in individual plant nuclei. This technology is particularly useful for understanding plant development, stress responses, and the molecular basis of traits such as drought tolerance and disease resistance.
Researchers have applied snRNA-seq to explore the gene expression landscapes of model plants like Arabidopsis and economically important crops such as rice and maize. These studies have identified key regulatory networks and genetic variants associated with desirable agronomic traits, facilitating crop improvement and sustainable agriculture.
snRNA-seq Can Be Utilized in A Variety Of Applications
snRNA-seq is revolutionizing our understanding of biological systems by providing a high-resolution view of cellular heterogeneity and gene expression dynamics. Its applications span a wide range of fields, from neuroscience and oncology to immunology and plant biology. With its inclusion, snRNA-seq offers new insights into complex biological processes and drives innovation in research and medicine.
As snRNA-seq technology continues to advance, its impact on science and healthcare is poised to grow. This could unlock new possibilities for precision medicine and sustainable agriculture. By harnessing the power of snRNA-seq, researchers are paving the way for breakthroughs that can transform our understanding of life at the molecular level.
If you have found this article interesting, refer to our blog page for more insights into RNA sequencing. Our blog features articles that delve into how RNA sequencing can enhance single-cell sequencing and its ability to categorize different types of cells.