Introduction
DeNovix DS-11 Series instruments use an automatic single-point correction for most applications. The correction subtracts the absorbance value at a specific wavelength from all wavelengths across the sample spectrum. The correction is used to account for the effect of instrument noise and light-scattering particulates in the sample that can cause an offset in the overall sample absorbance.
This technical note describes how baseline corrections are handled in various DS-11 apps and provides helpful hints on selecting baseline wavelengths for custom dyes and methods.
Potential Baseline Offset
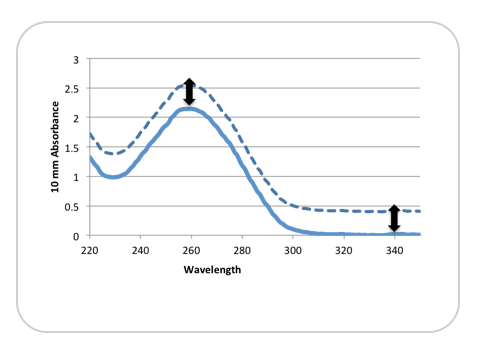
Figure 1 illustrates how an incorrect absorbance reading can be obtained from a non-baseline corrected sample.
The dotted line represents a sample exhibiting a significant shift from the baseline at 340 nm. The solid line represents the same sample that has been baseline corrected. For this example, the reported absorbance at 260 nm—and therefore, the calculated concentration—would be about 20% higher than expected.
Baseline Wavelength Selection
The optimal wavelength to use for baseline correction is a wavelength at which there is no absorbance attributed to the sample buffer or the molecule of interest. Most methods for standard absorbance based assays found in scientific literature will include the wavelength used as for baseline correction.
A general recommendation would be to use 340 nm for methods that use UV only wavelength ranges (190 – 350 nm) and 750 nm for methods using ranges that extend to 750 nm or greater.
Assessing a Baseline Correction Wavelength
Nucleic Acids and Protein Apps
Baseline correction for nucleic acid and protein applications have been traditionally performed at 320 nm. However, with the advent of microvolume spectrophotometers capable of reading highly concentrated samples, the correction is now routinely done using 340 nm.
The DeNovix dsDNA, ssDNA, RNA, Protein A280 and the Peptide apps all use 340 nm as the default baseline correction wavelength. Users may change the selection by using the Baseline Correction feature, which is accessed from the app Run screen Overflow menu.
Microarray and Labeled Proteins Apps
The software anchors the visual spectrum, displayed in both the Microarray and Labeled Protein apps, using a baseline of 750 nm for all measurements. A 340 nm bichromatic normalization is applied for the nucleic acid or protein analysis wavelengths (260 nm or 280 nm respectively), and a sloping baseline between 400 and 750 nm is applied to the dye-specific analysis wavelength dye concentration calculations.
When adding a new dye, it is important to empirically determine the appropriate baseline correction wavelength. User-defined options for Microarray and Labeled Protein apps are enabled between 750 and 840 nm. Consider using a baseline correction of 800 nm or greater for dyes with absorbance maxima greater than 700 nm.
The new selection will be used for both the visual display and the sloping dye baseline. Once a baseline correction wavelength is selected, it is applied to all subsequent measurements and is not dye specific. When changing dyes, always confirm that the proper baseline correction is applied.
The wavelength currently in use is displayed in small print at the bottom the Run screen.
Colorimetrics App
The default baseline correction wavelengths are preconfigured for each of the included assays types and may not be changed.
UV-Vis App
The default baseline correction wavelength for the UV-Vis app is 750 nm. However, it is suggested that the optimal wavelength be empirically determined for each sample type. Use the Baseline Correction feature, which is accessed from the Overflow menu to change selections.
Custom Method Apps
A baseline correction wavelength is required for the Standard Curve app methods but not for Formula Methods. The optimal baseline correction wavelength should be empirically determined for each method and should take into account the sample type and reagents to be measured.
A general recommendation would be to use 340 nm for UV only wavelength ranges and 750 nm for methods including the Vis wavelength ranges.
OD600 App
Microbial cell culture OD 600 values are measurements of light scattering. The DS-11 software does not use a baseline normalization unless specified as a user selection.
Kinetics App (DS-11+, DS-8X+, DS-7+, DS-C)
A baseline correction wavelength is required for all Kinetics app methods. As in all custom methods, the optimal baseline correction wavelength should be empirically determined.
28-MAR-2025