Introduction
The ability to measure microliter volumes of samples via UV-Vis spectrophotometry has been a revolution in molecular biology labs over the last decade. Understanding best practice for preparing and measuring your samples when using microvolume spectrophotometers will avoid sources of error that negatively affect the accuracy of results and impact the downstream analysis of a sample.
DeNovix Inc. was established by the original innovators in the field of microvolume spectrophotometry. DeNovix scientists bring decades of combined experience in absorbance and fluorescence analysis and present here a best practice guide to avoid common errors and ensure precise and accurate results.
1. The Importance of Sample Preparation
Ensure that sample isolation protocols are optimized and that the samples are purified prior to measurement. Keep in mind that a UV-Vis spec does not discriminate between the sample of interest and contaminants that also absorb at the same wavelength. All nucleic acids exhibit a peak at or around 260 nm. Degraded or single-stranded DNA in a double-stranded DNA sample solution will contribute to the total absorbance at 260 nm, resulting in the over estimation of the reported concentration.
Monitoring the measured absorbance ratios will provide an insight into the purity of the sample and identify potential issues in sample preparation. Pure DNA and RNA preparations have A260/A280 ratios of 1.8 and 2.0 respectively and an A260/230 ratio between 2 and 2.2. Deviation from these can indicate carryover of protein for the former or carbohydrate, guanidine, phenol or glycogen for the latter ratio.
Purity ratios may also be impacted when an inappropriate solution is used as a blank or if samples are very dilute and close to the lower detection limit.
2. Sample Surface Cleaning
Ensuring that both top and bottom measurement surfaces are clean prior to loading blanks or samples is crucial to avoiding problems with subsequent readings.
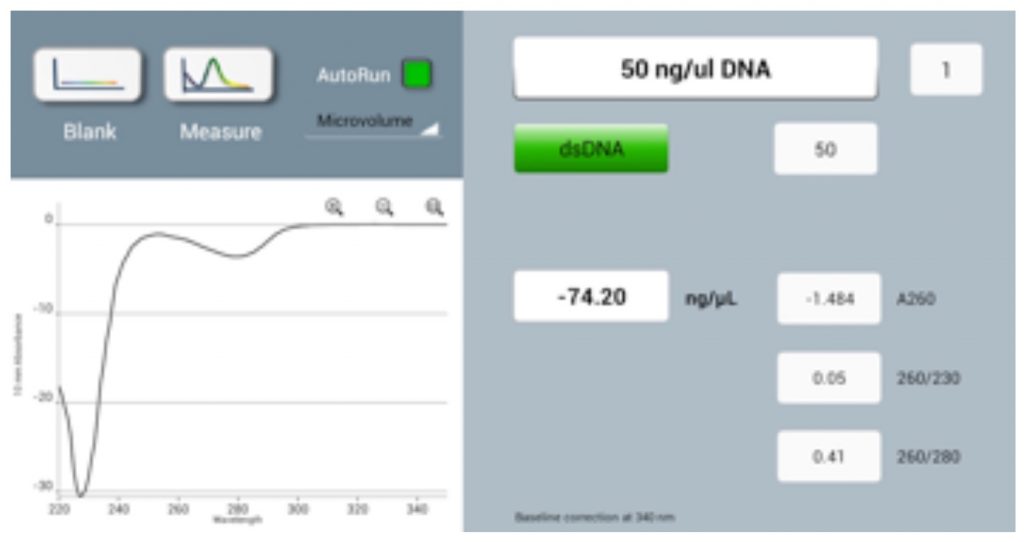
Performing a blank measurement on a dirty sampling surface (either top or bottom) will result in erroneous absorbance values, such as negative spectra (Figure 1) or sample concentrations measured as lower than the actual values.
Over a period of time, detergents or alcohols in the buffers of some samples can lead to the unconditioning of the measurement surface, causing the sample to lie flat rather than bead up on the pedestal.
If any of the issues described are observed, it is important to follow the manufacturer’s recommended cleaning protocol before re-blanking and continuing to measure samples.
3. Blank Measurements and Buffers
An appropriate Blank is a prerequisite for accurate concentration measurements. Use the same buffer as the sample is suspended in for the blank reading and ensure that the measurement surfaces are clean prior to loading. Seeing negative spectra when running samples is a strong indicator that either the sample pedestals were dirty or a sample was measured as a blank in error.
When selecting buffers, avoid those that contain components that absorb strongly at the wavelength of interest. For example, proteins suspended in RIPA buffers, which have a high absorbance at 280 nm. When these buffers are required, colorimetric assays are recommended. The absorbance profile of buffers can be checked by blanking with water and running the buffer as a sample.
4. Measure samples within the limits of detection the spectrophotometer
All spectrophotometers have a detectable level of background noise contributed by the environment and electronic components. The Lower Detection Limit of an instrument can be defined as the lowest quantity of analyte that can be distinguished from the background noise.
A tolerance specification may be given as either a percentage value or as a concentration for a specific analyte. For example, where the tolerance specification is 2 ng/µL for dsDNA, a sample with a concentration of 4 ng/µL could range from 2 to 6 ng/µL due to the contribution from electrical noise (see Table 1).
Note that the DS-11 Series has an industry-leading lower detection limit for dsDNA of 0.75 ng/µL.
When working with samples at or near the lower detection limit, switching from microvolume to using the longer pathlength 1 cm cuvette mode, as on the DS-11+ UV-Vis Spectrophotometer, extends the acceptable lower measurement range 20-fold.
Table 1: Tolerance Specifications| Target Concentration (ng/μL dsDNA) | Tolerance Specification (ng/μL dsDNA)* | Maximum %Error |
|---|---|---|
| 1000 | +/- 10 | 1% |
| 200 | +/- 2 | 1.0% |
| 100 | +/- 2 | 2.0% |
| 10 | +/- 2 | 20% |
| 4 | +/- 2 | 50% |
| 2 | +/- 2 | 100% |
*Tolerance specifications stated are in a range typical for microvolume spectrophotometers.
5. Routine best practices
Summary
Being able to accurately measure the UV-Vis absorbance of samples in microliter volumes enables researchers to conserve their precious materials when performing quality control steps essential for downstream applications. Adopting these simple tips when quantifying nucleic acids or proteins will help deliver additional accuracy and confidence in your microvolume measurements.
Revised 19 Oct 2020