Materials
Procedure
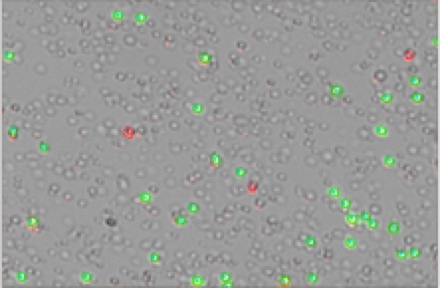
Cell counting and viability measurements are performed using the DeNovix Acridine Orange (AO) / Propidium Iodide (PI) Assay. AO is a nucleic acid-binding fluorophore that is cell membrane permeable and suitable for selective staining of nucleated PBMCs. PI is a nucleic acid-binding dye that is impermeable to live cells and suitable for staining dead, nucleated PBMCs. All live, nucleated cells fluoresce green due to AO. Dead, nucleated cells are stained with both AO and PI and fluoresce red. RBCs and debris are unstained and not counted.
Cell Size and Fluorescence Thresholds
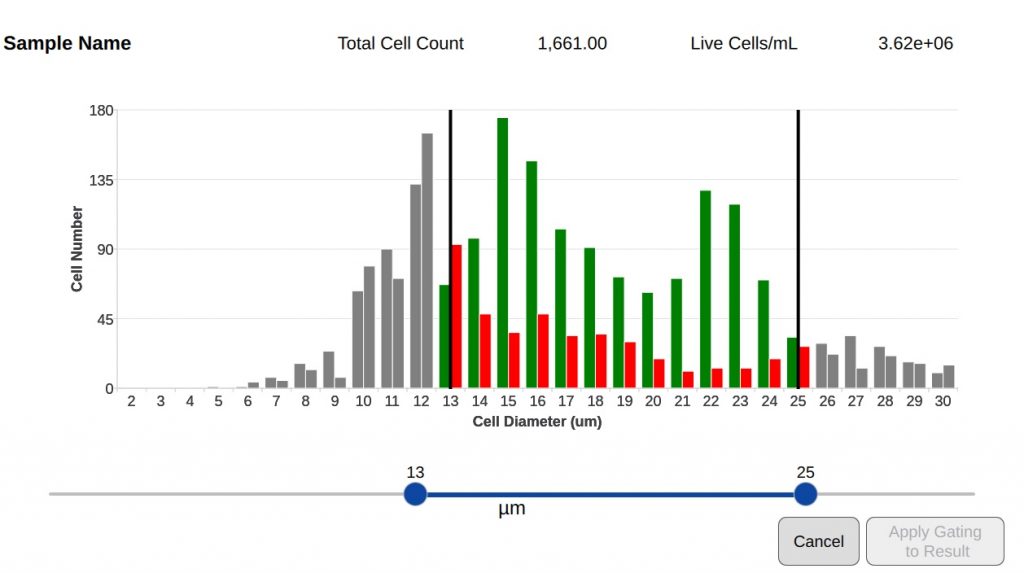
Setting an appropriate size range and fluorescence intensity for the cells of interest can exclude debris or alternative cell populations in a sample from analysis. Minimum and maximum cell diameters along with appropriate intensity thresholds can be defined and saved in protocols.
The min and max diameter can be dynamically altered using the cell size histogram once cells have been counted. The data is rapidly reanalyzed to take account of changed settings. For more advanced reanalysis, the Optimize Settings button allows the user to access and change all protocol settings on the current image and recount the sample.
Once a protocol has been optimized, the user can edit and save the original protocol with these new settings, allowing for accurate and rapid counts of future samples and standardization between different researchers.
Chamber Height
The CellDrop has a unique, adjustable height sample chamber that can range from 50 to 400 µm. Setting the height at 400 µm allows for a more accurate count of low density samples.
A chamber height of 50 µm allows a higher density sample to be counted without the need for further dilution. This feature gives the CellDrop the greatest dynamic range of any image based counter on the market. The chamber height can be selected from the protocols screen. The recommended concentration range at each chamber height is detailed in Table 1.
(µm) | Sample Volume (µL) | Min Cell Density (cells/mL) | Max Cell Density (cells/mL) |
|---|---|---|---|
| 400 | 40 | 7.0 x 102 | 1.0 x 105 |
| 100 | 10 | 5.0 x 104 | 1.0 x 107 |
| 50 | 5 | 1.0 x 10 7 | 2.5 x 107 |
Summary
Automated counting of PBMC samples on the CellDrop FLi removes operator variability from the process, speeds up the workflow and enables customizable reporting and data archiving. Dual fluorescence measurements using AO/PI allow the specific identification of live and dead PBMCs in the presence of large numbers of red blood cells, platelets and cellular debris.
While AO/PI are frequently used fluorophores for this application, the CellDrop is able to measure a wide range of common fluorophores. Contact our Applications Support Team at info@denovix.com to discuss specific assay requirements.
19-NOV-2024